Things will get better............u really do NOT understand.......u have ruined my fucking life.........i am NOT your fucking prostitute.......NOR am i a FUCKING spy.....
Genetic recombination
 |
 |
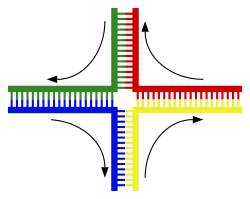
Structure of the Holliday junction intermediate in genetic recombination. The four separate DNA strands are coloured red, blue, green and yellow.[149]
A DNA helix usually does not interact with other segments of DNA, and in human cells, the different chromosomes even occupy separate areas in the nucleus called "chromosome territories".[150] This physical separation of different chromosomes is important for the ability of DNA to function as a stable repository for information, as one of the few times chromosomes interact is in chromosomal crossover which occurs during sexual reproduction, when genetic recombination occurs. Chromosomal crossover is when two DNA helices break, swap a section and then rejoin.
Recombination allows chromosomes to exchange genetic information and produces new combinations of genes, which increases the efficiency of natural selection and can be important in the rapid evolution of new proteins.[151] Genetic recombination can also be involved in DNA repair, particularly in the cell's response to double-strand breaks.[152]
The most common form of chromosomal crossover is homologous recombination, where the two chromosomes involved share very similar sequences. Non-homologous recombination can be damaging to cells, as it can produce chromosomal translocations and genetic abnormalities. The recombination reaction is catalyzed by enzymes known as recombinases, such as RAD51.[153] The first step in recombination is a double-stranded break caused by either an endonuclease or damage to the DNA.[154] A series of steps catalyzed in part by the recombinase then leads to joining of the two helices by at least one Holliday junction, in which a segment of a single strand in each helix is annealed to the complementary strand in the other helix. The Holliday junction is a tetrahedral junction structure that can be moved along the pair of chromosomes, swapping one strand for another. The recombination reaction is then halted by cleavage of the junction and re-ligation of the released DNA.[155] Only strands of like polarity exchange DNA during recombination. There are two types of cleavage: east-west cleavage and north-south cleavage. The north-south cleavage nicks both strands of DNA, while the east-west cleavage has one strand of DNA intact. The formation of a Holliday junction during recombination makes it possible for genetic diversity, genes to exchange on chromosomes, and expression of wild-type viral genomes.
Evolution
DNA contains the genetic information that allows all forms of life to function, grow and reproduce. However, it is unclear how long in the 4-billion-year history of life DNA has performed this function, as it has been proposed that the earliest forms of life may have used RNA as their genetic material.[156][157] RNA may have acted as the central part of early cell metabolism as it can both transmit genetic information and carry out catalysis as part of ribozymes.[158] This ancient RNA world where nucleic acid would have been used for both catalysis and genetics may have influenced the evolution of the current genetic code based on four nucleotide bases. This would occur, since the number of different bases in such an organism is a trade-off between a small number of bases increasing replication accuracy and a large number of bases increasing the catalytic efficiency of ribozymes.[159] However, there is no direct evidence of ancient genetic systems, as recovery of DNA from most fossils is impossible because DNA survives in the environment for less than one million years, and slowly degrades into short fragments in solution.[160] Claims for older DNA have been made, most notably a report of the isolation of a viable bacterium from a salt crystal 250 million years old,[161] but these claims are controversial.[162][163]
Building blocks of DNA (adenine, guanine, and related organic molecules) may have been formed extraterrestrially in outer space.[164][165][166] Complex DNA and RNA organic compounds of life, including uracil, cytosine, and thymine, have also been formed in the laboratory under conditions mimicking those found in outer space, using starting chemicals, such as pyrimidine, found in meteorites. Pyrimidine, like polycyclic aromatic hydrocarbons (PAHs), the most carbon-rich chemical found in the universe, may have been formed in red giants or in interstellar cosmic dust and gas clouds.[167]
Uses in technology
Genetic engineering
Methods have been developed to purify DNA from organisms, such as phenol-chloroform extraction, and to manipulate it in the laboratory, such as restriction digests and the polymerase chain reaction. Modern biology and biochemistry make intensive use of these techniques in recombinant DNA technology. Recombinant DNA is a man-made DNA sequence that has been assembled from other DNA sequences. They can be transformed into organisms in the form of plasmids or in the appropriate format, by using a viral vector.[168] The genetically modified organisms produced can be used to produce products such as recombinant proteins, used in medical research,[169] or be grown in agriculture.[170][171]
DNA profiling
Forensic scientists can use DNA in blood, semen, skin, saliva or hair found at a crime scene to identify a matching DNA of an individual, such as a perpetrator.[172] This process is formally termed DNA profiling, also called DNA fingerprinting. In DNA profiling, the lengths of variable sections of repetitive DNA, such as short tandem repeats and minisatellites, are compared between people. This method is usually an extremely reliable technique for identifying a matching DNA.[173] However, identification can be complicated if the scene is contaminated with DNA from several people.[174] DNA profiling was developed in 1984 by British geneticist Sir Alec Jeffreys,[175] and first used in forensic science to convict Colin Pitchfork in the 1988 Enderby murders case.[176]
The development of forensic science and the ability to now obtain genetic matching on minute samples of blood, skin, saliva, or hair has led to re-examining many cases. Evidence can now be uncovered that was scientifically impossible at the time of the original examination. Combined with the removal of the double jeopardy law in some places, this can allow cases to be reopened where prior trials have failed to produce sufficient evidence to convince a jury. People charged with serious crimes may be required to provide a sample of DNA for matching purposes. The most obvious defense to DNA matches obtained forensically is to claim that cross-contamination of evidence has occurred. This has resulted in meticulous strict handling procedures with new cases of serious crime.
DNA profiling is also used successfully to positively identify victims of mass casualty incidents,[177] bodies or body parts in serious accidents, and individual victims in mass war graves, via matching to family members.
DNA profiling is also used in DNA paternity testing to determine if someone is the biological parent or grandparent of a child with the probability of parentage is typically 99.99% when the alleged parent is biologically related to the child. Normal DNA sequencing methods happen after birth, but there are new methods to test paternity while a mother is still pregnant.[178]
DNA enzymes or catalytic DNA
Deoxyribozymes, also called DNAzymes or catalytic DNA, were first discovered in 1994.[179] They are mostly single stranded DNA sequences isolated from a large pool of random DNA sequences through a combinatorial approach called in vitro selection or systematic evolution of ligands by exponential enrichment (SELEX). DNAzymes catalyze variety of chemical reactions including RNA-DNA cleavage, RNA-DNA ligation, amino acids phosphorylation-dephosphorylation, carbon-carbon bond formation, and etc. DNAzymes can enhance catalytic rate of chemical reactions up to 100,000,000,000-fold over the uncatalyzed reaction.[180] The most extensively studied class of DNAzymes is RNA-cleaving types which have been used to detect different metal ions and designing therapeutic agents. Several metal-specific DNAzymes have been reported including the GR-5 DNAzyme (lead-specific),[179] the CA1-3 DNAzymes (copper-specific),[181] the 39E DNAzyme (uranyl-specific) and the NaA43 DNAzyme (sodium-specific).[182] The NaA43 DNAzyme, which is reported to be more than 10,000-fold selective for sodium over other metal ions, was used to make a real-time sodium sensor in cells.
Bioinformatics
Bioinformatics involves the development of techniques to store, data mine, search and manipulate biological data, including DNA nucleic acid sequence data. These have led to widely applied advances in computer science, especially string searching algorithms, machine learning, and database theory.[183] String searching or matching algorithms, which find an occurrence of a sequence of letters inside a larger sequence of letters, were developed to search for specific sequences of nucleotides.[184] The DNA sequence may be aligned with other DNA sequences to identify homologous sequences and locate the specific mutations that make them distinct. These techniques, especially multiple sequence alignment, are used in studying phylogenetic relationships and protein function.[185] Data sets representing entire genomes' worth of DNA sequences, such as those produced by the Human Genome Project, are difficult to use without the annotations that identify the locations of genes and regulatory elements on each chromosome. Regions of DNA sequence that have the characteristic patterns associated with protein- or RNA-coding genes can be identified by gene finding algorithms, which allow researchers to predict the presence of particular gene products and their possible functions in an organism even before they have been isolated experimentally.[186] Entire genomes may also be compared, which can shed light on the evolutionary history of particular organism and permit the examination of complex evolutionary events.
DNA nanotechnology
DNA nanotechnology uses the unique molecular recognition properties of DNA and other nucleic acids to create self-assembling branched DNA complexes with useful properties.[187] DNA is thus used as a structural material rather than as a carrier of biological information. This has led to the creation of two-dimensional periodic lattices (both tile-based and using the DNA origami method) and three-dimensional structures in the shapes of polyhedra.[188] Nanomechanical devices and algorithmic self-assembly have also been demonstrated,[189] and these DNA structures have been used to template the arrangement of other molecules such as gold nanoparticles and streptavidin proteins.[190]
History and anthropology
Because DNA collects mutations over time, which are then inherited, it contains historical information, and, by comparing DNA sequences, geneticists can infer the evolutionary history of organisms, their phylogeny.[191] This field of phylogenetics is a powerful tool in evolutionary biology. If DNA sequences within a species are compared, population geneticists can learn the history of particular populations. This can be used in studies ranging from ecological genetics to anthropology.
Information storage
DNA as a storage device for information has enormous potential since it has much higher storage density compared to electronic devices. However high costs, extremely slow read and write times (memory latency), and insufficient reliability has prevented its practical use.[192][193]
History
DNA was first isolated by the Swiss physician Friedrich Miescher who, in 1869, discovered a microscopic substance in the pus of discarded surgical bandages. As it resided in the nuclei of cells, he called it "nuclein".[194][195] In 1878, Albrecht Kossel isolated the non-protein component of "nuclein", nucleic acid, and later isolated its five primary nucleobases.[196][197]
In 1909, Phoebus Levene identified the base, sugar, and phosphate nucleotide unit of the RNA (then named "yeast nucleic acid").[198][199][200] In 1929, Levene identified deoxyribose sugar in "thymus nucleic acid" (DNA).[201] Levene suggested that DNA consisted of a string of four nucleotide units linked together through the phosphate groups ("tetranucleotide hypothesis"). Levene thought the chain was short and the bases repeated in a fixed order. In 1927, Nikolai Koltsov proposed that inherited traits would be inherited via a "giant hereditary molecule" made up of "two mirror strands that would replicate in a semi-conservative fashion using each strand as a template".[202][203] In 1928, Frederick Griffith in his experiment discovered that traits of the "smooth" form of Pneumococcus could be transferred to the "rough" form of the same bacteria by mixing killed "smooth" bacteria with the live "rough" form.[204][205] This system provided the first clear suggestion that DNA carries genetic information.
In 1933, while studying virgin sea urchin eggs, Jean Brachet suggested that DNA is found in the cell nucleus and that RNA is present exclusively in the cytoplasm. At the time, "yeast nucleic acid" (RNA) was thought to occur only in plants, while "thymus nucleic acid" (DNA) only in animals. The latter was thought to be a tetramer, with the function of buffering cellular pH.[206][207]
In 1937, William Astbury produced the first X-ray diffraction patterns that showed that DNA had a regular structure.[208]
In 1943, Oswald Avery, along with co-workers Colin MacLeod and Maclyn McCarty, identified DNA as the transforming principle, supporting Griffith's suggestion (Avery–MacLeod–McCarty experiment).[209] DNA's role in heredity was confirmed in 1952 when Alfred Hershey and Martha Chase in the Hershey–Chase experiment showed that DNA is the genetic material of the enterobacteria phage T2.[210]
Late in 1951, Francis Crick started working with James Watson at the Cavendish Laboratory within the University of Cambridge. In February 1953, Linus Pauling and Robert Corey proposed a model for nucleic acids containing three intertwined chains, with the phosphates near the axis, and the bases on the outside.[211] In May 1952, Raymond Gosling a graduate student working under the supervision of Rosalind Franklin took an X-ray diffraction image, labeled as "Photo 51",[212] at high hydration levels of DNA. This photo was given to Watson and Crick by Maurice Wilkins and was critical to their obtaining the correct structure of DNA. Franklin told Crick and Watson that the backbones had to be on the outside. Before then, Linus Pauling, and Watson and Crick, had erroneous models with the chains inside and the bases pointing outwards. Her identification of the space group for DNA crystals revealed to Crick that the two DNA strands were antiparallel.[213]



No comments:
Post a Comment